- The work, published in Nature Genetics, represents an important advance in our understanding of gene regulation and reveals a new layer of complexity that needs to be studied to properly interpret genomics and gene expression in the future.
- An example of how risky fundamental science with innovative approaches leads to surprising and important advances in knowledge.
Barcelona, January 15th 2018.- Scientists have long been reading the code of life – the genome –, as a sequence of letters but now researchers have also started exploring its three-dimensional organisation. In a paper published in Nature Genetics, an interdisciplinary research team of scientists from the Centre for Genomic Regulation (CRG) – including a Centro Nacional de Análisis Genómico (CNAG-CRG) group – in Barcelona, Spain, shows that the three-dimensional organisation of the genome plays a key role in gene expression and consequently in determining cell fate.
It all began with the 4D Genome project, an ambitious and innovative research initiative funded by a European Research Council Synergy grant aimed at understanding how the spatial organisation of the genome contributes to decisions made by cells. What scientists wanted to find out was whether genome architecture plays an active function or whether it is a mere side effect of the genome's activity. The model that the 4D Genome team used was cell reprogramming, a process that allows the scientists to revert white blood cells back to a state of pluripotency whereby these cells can differentiate into any other cell type. Proteins that control gene activity (known as transcription factors) play a key role in this process, which the 4D Genome team studied in great detail, assessing how they induce changes in gene expression, modifications of chromatin (the structure around which DNA is wrapped), and changes in the 3D organisation of the genome.
Understanding how stem cells are formed and how they can convert into different cell types is a major challenge in modern biology. It has been known since the fifties that all specialized cells in the body contain the same genome. So, what then distinguishes one cell type from another? The answer is that different cells read the information contained in different parts of the genome. The machinery required to achieve this selective information recall resides primarily in so called ‘transcription factors’, molecules that switch genes on or off. Many other molecules are also involved in this process, including epigenetic regulators that help to densely pack the genome into the nucleus of a cell, or locally unpack it to allow gene expression. The scientists used a reprogramming method discovered by CRG senior researcher Thomas Graf and his team. With this precision tool in hand, they were able to study the dynamics of genome organization by comparing the changes in genome architecture and transcription at different times during reprogramming.
“We expected that transcription factors would first switch on certain genes, which then afterwards may force a re-organisation of the 3D structure of the chromosome. Surprisingly, what we found is that in a large proportion of the genome, the transcription factors were actually promoting this re-organisation before the genes were switched on,” explains Ralph Stadhouders, co-first author of the paper together with computational biologist Enrique Vidal.
“Our paper shows that transcription factors may play a completely new role in cell reprogramming: they do not only switch genes on and off, but also promote the architectural changes necessary to modify gene expression,” states Thomas Graf, senior CRG group leader and lead investigator of this study. “It may have relevant implications for researchers worldwide studying gene regulation in general since we know now that the large-scale organization of the genome cannot be ignored. It also raises new questions about how these large changes can be brought about: possibly new mechanisms, perhaps even new machineries that are necessary to change certain areas of the genome” reflects Graf.
The perfect combination: risky + interdisciplinary fundamental science
The findings reported in Nature Genetics indicate that genome architecture has important informational value for controlling gene expression during cell reprogramming, and is thus required for the specialized functions of a cell. "We are only scratching the surface of what could be critical as yet unrecognized mechanisms by which cells regulate gene expression" says co-principal investigator and ICREA research professor at CNAG-CRG, Marc A. Martí-Renom. “The new discovery might also be fundamental for development and for some development-related diseases and cancer,” he concludes.
Again, this is an example on how risky science with innovative and interdisciplinary approaches leads to surprising and relevant results to advance knowledge. In this case, four laboratories at the Centre for Genomic Regulation (including a CNAG-CRG one) took full advantage of their experience and diverse capabilities to address fundamental questions in genome biology with the support of a 13M € ERC Synergy grant*. “Promoting and supporting fundamental research is crucial to advance knowledge. We are proud that our project is already providing the first answers to the burning question about how genome 3D organisation shapes genome regulation,” agree the four group leaders participating in the 4D Genome project: Thomas Graf, Marc A. Martí-Renom, Guillaume Filion, and Miguel Beato.
*ERC Synergy grants support scientific projects carried out by different interdisciplinary groups of researchers tackling issues at the forefront of knowledge, new areas of research, and novel methods and techniques.
Work of reference
Ralph Stadhouders, Enrique Vidal, François Serra, Bruno Di Stefano, François Le Dily, Javier Quilez, Antonio Gomez, Samuel Collombet, Clara Berenguer, Yasmina Cuartero, Jochen Hecht, Guillaume J. Filion, Miguel Beato, Marc A. Marti-Renom and Thomas Graf. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nature Genetics (2018). DOI: 10.1038/s41588-017-0030-7.
Funding information
This work was supported by the European Research Council under the 7th Framework Program FP7/2007-2013 (ERC Synergy Grant 4D Genome, grant agreement 609989) and the Ministerio de Educacion y Ciencia, SAF.2012-37167. R.S. was supported by an EMBO Long-term Fellowship (ALTF 12012014) and a Marie Curie Individual Fellowship (H2020 MSCA-IF-2014). We also acknowledge support from ‘Centro de Excelencia Severo Ochoa 2013-2017’ (SEV-2012-0208) and AGAUR to the CRG.
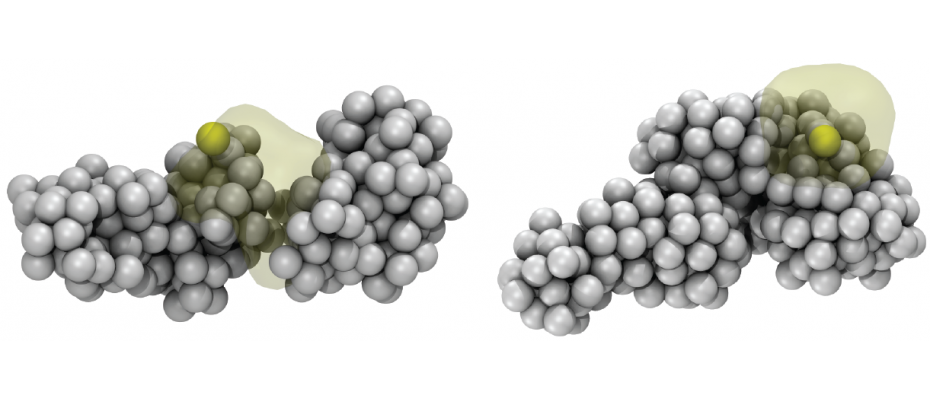
Image Caption
Architectural changes in chromosome 13 around the pluripotency stem cell gene Sox2 (yellow ball) during reprogramming of blood cells (left) into induced pluripotent cells (right). (Credit: Marco Di Stefano, CNAG-CRG).










